Companies boasting about their diapers’ comfort, mobility, and, most importantly, absorbency, represent the hallmark of any respected diaper brand. If you have ever seen a diaper commercial, you have seen toddlers sporting their amazingly elastic and stretchable bottom-wear. Every advertisement has a scene where the absorbent power of the diaper in question is put to the test.
The pivotal feature of diapers responsible for this impressive feat is a synthetic molecule called sodium polyacrylate, which can absorb over 1,000 times its mass in water. As a cost-effective and hyper-absorbent substance, sodium polyacrylate is also used in sanitary napkins, anti-flood bags, medical bandages, and much more. It is actually rather common to see a successful product form after a radically unsuccessful original purpose, such as lysol, play-doh, and bubble wrap. However, there is seldom a product with multiple successful applications in many different markets. Recently, sodium polyacrylate checked off another venture as it found its way into the world of neuroscience through its emerging implementation in microscope technology.
Microscope technology has undergone rapid innovation since its inception, with scientists now observing single atoms (one billionth of a meter in length) using powerful electron microscopes. Although electron microscopes answer many important questions, typical labs or facilities do not have upwards of $25 million to acquire such devices, so they often make use of state-of-the-art optical microscopes instead. However, optical microscope technology has reached the limit of its capabilities. Physicists have known for decades that there is a physical constraint when making observations with visible light, which is about 200 nanometers (nm), or 500 times thinner than the width of human hair. This quandary has led to the development of many innovative super-resolution imaging techniques that have attempted to mitigate light’s diffraction limit. These techniques, such as STORM or STED microscopy, can bring the optical constraint of microscopes down from 200 nm to around 25 nm by using physical principles to counteract the wavelength of light [1]. Recently, however, a new imaging technique has challenged the conventional approach of microscope visualization by making the physical sample larger instead of attempting to magnify it further. Sodium polyacrylate, the foundation of modern diaper technology, is the key to this imaging revolution.
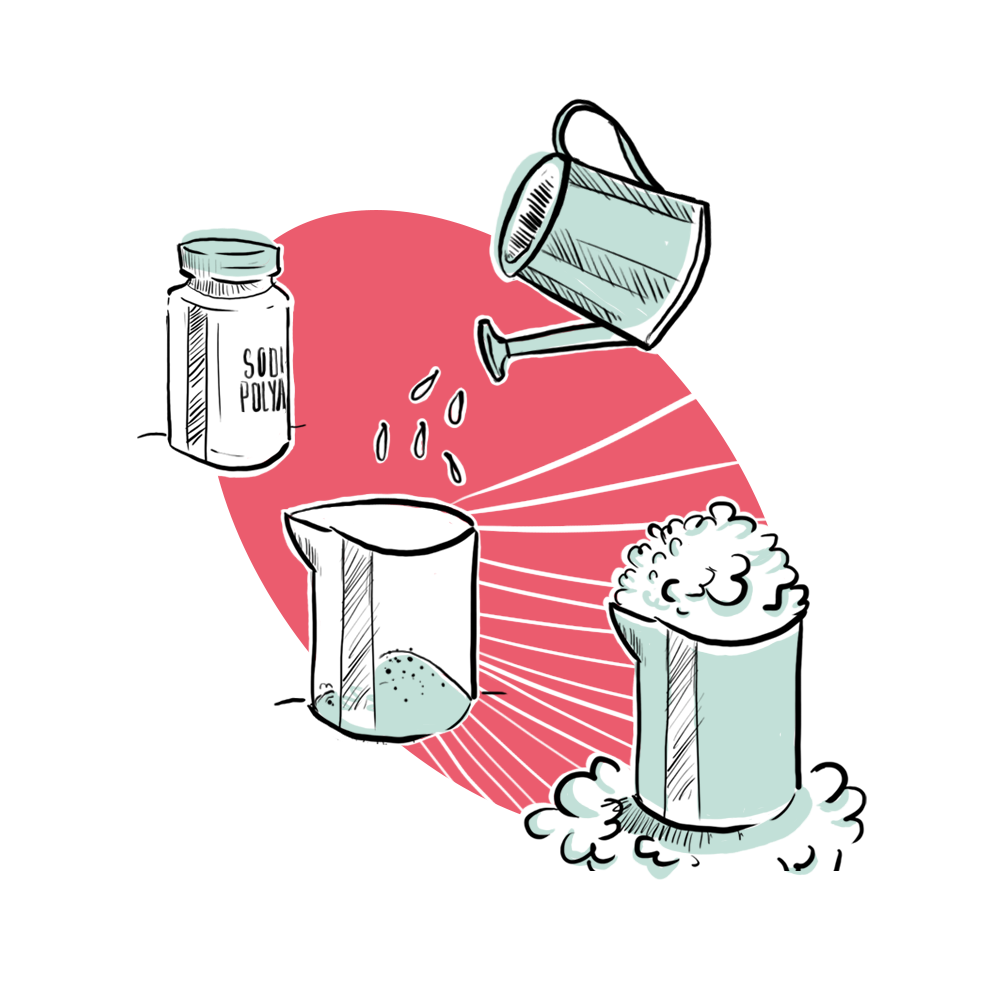
In 2015, Dr. Ed Boyden and his team at MIT first proposed a novel cellular imaging technique called expansion microscopy (ExM) [1]. ExM works by incorporating certain biomolecules into a tissue sample that act as anchors for a web of sodium polyacrylate polymers. The polymer is attached to the anchors in the tissue sample every few nanometers, so structures and molecules that are extremely complex and close together can be separated for better visualization when the swelling occurs.
When water is added, the swellable polymer expands the tissue sample 100-fold or more in volume while maintaining the structural integrity of and dimensional relationships within the tissue sample. This yields a more magnified, 70 nm or higher resolution depiction of the intricate connections and structures of the tissue sample in question [1]. Better visualization is particularly important in neuroscience because many molecules and wirings in neural circuits are not well understood, particularly when they are not functioning properly. Mapping and visualizing neurons in these neural circuits have posed a great challenge for neuroscientists, and ExMis attempting to alleviate this conundrum [2].
ExM is said to be faster, easier to use, and less expensive than the aforementioned super-resolution microscopy methods that often require more money, extensive knowledge, and training. Synapses can be expanded in a fashion that allows the imaging of scaffolding proteins, neurotransmitter receptors, and other densely packed synaptic components [3]. ExM has been successfully applied to mouse brain, mouse spinal cord, monkey brain, human sections, and many other tissue sections, making it a widely versatile technology [4].
One such benefit of ExM has been demonstrated through experiments on radial glia, which are specialized cells most prominently found during early development that are capable of generating neurons; the tracing of radial glia processes is crucial for developmental neuroscientists. In zebrafish, ExM enabled the tracing of radial glia processes that were not previously resolvable with diffraction-limited microscopy alone. Additionally, it provided better resolution for the molecular differences between densely-packed neural synapses and gave better characterization to shapes of cytoskeletal proteins [5]. The cytoskeleton of a cell is a complex set of polymers made of interlinking proteins that give a cell its structural support and shape and is integral to how cells divide and proliferate. Identifying new proteins or developing a better understanding of the function of existing proteins in this process is essential to uncovering the unknown parts of cell reproduction. These neurological discoveries are in large part due to ExM and its novel use of sodium polyacrylate, a molecule most commonly used to clean up after infants.
This technology has only been in existence for about three years, but it is rapidly flourishing, improving, and being incorporated into an increasing number of laboratories. In 2016, an ExM variation, called proExM, enabled the use of more commonly used fluorescent labels, as the original ExM procedure previously required specialized fluorescent labels which often cost more money [6]. Fluorescent labels are chemical tags that scientists can selectively attach to proteins or other structures and emit certain colors depending on the tag that is used. These labels allow scientists to color samples when viewed under specific microscope conditions in order to distinguish structures from each other and achieve better visualization [6]. Beyond lowering the cost of the original ExM protocol, proExM has proven useful when characterizing astrocytic gap junctions near blood vessels in human epileptic brain specimens [7]. Astrocytes are a type of glial cell, which surround neural cells and provide support for and insulation between neurons to help them with conductivity and separation. Astrocytes’ main functions are redistributing ions and water, glucose metabolism, and nerve cell communication; deficits in all of these areas have been associated with epilepsy [8]. Simply being able to visualize compact and complex structures in a new expanded way helps develop interesting insights and further questions for neuroscientists.
In 2017, following the introduction of proExM, another ExM variant surfaced named iExM [9]. After the initial swelling in the original ExM procedure, a second swellable mesh was formed in the newly opened spaces from the first expansion and was expanded again. This brought the optical resolution down from the diffraction limit of 200 nm to an impressive 25 nm, which is commensurate with the aforementioned pre-ExM super-resolution imaging techniques. Not only does iExM give similar resolution results, but the technology also creates large amounts of space and separation between densely packed, intricate structures that would be hard to interpret otherwise. For example, neural synapses contain highly complicated interconnected networks of neurotransmitters and proteins working in tandem. iExM has allowed neuroscientists to visualize these synapses and, in mouse brains, visualize the detailed architecture of dendrites—short, branched extensions of neural cells [9].
One important spin-off that arose just three months after iExM is called ExPath, and it has implications that reach into the medical field [10]. ExPath is a procedure that can convert clinical tissue samples into ExM-compatible samples. By doing this, pathologists can now visualize structures that previously required expensive imaging or were simply too ambiguous to definitively diagnose. For example, kidney minimal change disease, which causes an excess loss of protein through urine, used to require confirmation via expensive electron microscopes but now can be optically diagnosed. Additionally, pathologists often disagreed when classifying possibly cancerous breast lesions, but ExPath allows for a clearer and more accurate differential diagnosis of biopsied tissue [10]. This ExM-based procedure may enable routine use of light microscopes in pathology and clinical research that previously would have required the use of costly electron microscope imaging.
As recently as September 2018, an ExM derived technique called X10 not only claims that its improved gel recipe gives X10 similar 25 nm resolution but that it is also the cheapest and easiest procedure that should be usable in all laboratories irrespective of specialized machinery or technical knowledge [11]. X10 is much too young to have given any scientific results thus far, but it demonstrates how excited researchers are to pick up ExM and try to improve it. Since its inception, four ExM derivatives have been developed, leading to many successful and novel findings. One by one, these findings will help unlock and unpack the mystery of the human brain one imaged synapse and protein at a time.
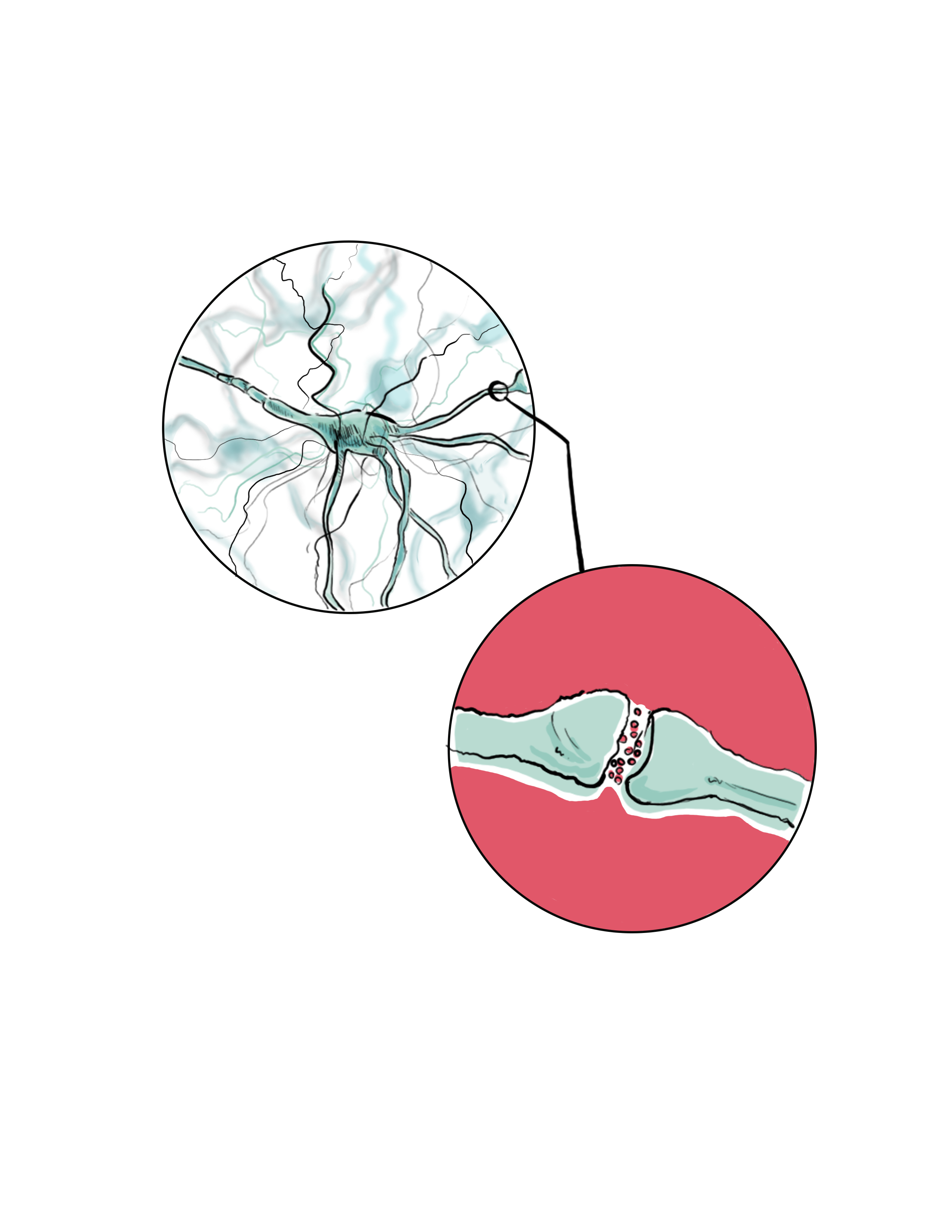
The human brain is one of the most compact, interconnected super-structures known to man, with a minimum of 100 trillion synapses. This is 1000 times more connections than the estimated number of stars in the Milky Way [12]. With such density and intricacy, it is no wonder that in order to achieve an accurate depiction of these inner workings, it is important to be able to separate them while maintaining their dimensional associations and integrity. Expansion microscopy has given scientists in clinical research, pathology, and especially those in neuroscience a simpler, faster, and better resolution approach to overcome the diffraction-limit of optical microscopes. For a technology that is only as old as the iPhone 6S, there has been an impressive amount of positive reception for and innovation done to this procedure. It should be noted, however, that this technology is still developing and is not a clear-cut, guaranteed success. Like any new invention, logistical problems, bugs, or kinks must always be ironed out over time. With that caveat in mind, the potential impact of ExM, especially in possible combinations with existing super-resolution microscopy methods, has an optimistic outlook. It will be exciting to see how this technology will expand the work of scientists who might have experienced economic constraints without the widespread accessibility of expansion microscopy.
References
- Chen, F., Tillberg, P. W., & Boyden, E. S. (2015). Optical imaging. Expansion microscopy. Science (New York, N.Y.) , 347 (6221), 543–548. https://doi.org/10.1126/science.1260088
- Karagiannis, E. D., & Boyden, E. S. (2018). Expansion microscopy: development and neuroscience applications. Current Opinion in Neurobiology , 50 , 56–63. https://doi.org/10.1016/j.conb.2017.12.012
- Gao, R., Asano, S. M., & Boyden, E. S. (2017). Q&A: Expansion microscopy. BMC Biology , 15 (1), 50. https://doi.org/10.1186/s12915-017-0393-3
- Cho, I., Seo, J. Y., & Chang, J. (2018). Expansion microscopy. Journal of Microscopy . https://doi.org/10.1111/jmi.12712
- Freifeld, L., Odstrcil, I., Förster, D., Ramirez, A., Gagnon, J. A., Randlett, O., … Boyden, E. S. (2017). Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proceedings of the National Academy of Sciences of the United States of America , 114 (50), E10799–E10808. https://doi.org/10.1073/pnas.1706281114
- Tillberg, P. W., Chen, F., Piatkevich, K. D., Zhao, Y., Yu, C.-C. J., English, B. P., … Boyden, E. S. (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology , 34 (9), 987–992. https://doi.org/10.1038/nbt.3625
- Deshpande, T., Li, T., Herde, M. K., Becker, A., Vatter, H., Schwarz, M. K., … Bedner, P. (2017). Subcellular reorganization and altered phosphorylation of the astrocytic gap junction protein connexin43 in human and experimental temporal lobe epilepsy. Glia , 65 (11), 1809–1820. https://doi.org/10.1002/glia.23196
- Heuser, K., Szokol, K., & Taubøll, E. (2014). The role of glial cells in epilepsy. Tidsskrift for Den Norske Laegeforening: Tidsskrift for Praktisk Medicin, Ny Raekke , 134 (1), 37–41. https://doi.org/10.4045/tidsskr.12.1344
- Chang, J.-B., Chen, F., Yoon, Y.-G., Jung, E. E., Babcock, H., Kang, J. S., … Boyden, E. S. (2017). Iterative expansion microscopy. Nature Methods , 14 (6), 593–599. https://doi.org/10.1038/nmeth.4261
- Zhao, Y., Bucur, O., Irshad, H., Chen, F., Weins, A., Stancu, A. L., … Boyden, E. S. (2017). Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nature Biotechnology , 35 (8), 757–764. https://doi.org/10.1038/nbt.3892
- Truckenbrodt, S., Maidorn, M., Crzan, D., Wildhagen, H., Kabatas, S., & Rizzoli, S. O. (2018). X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Reports , 19 (9). https://doi.org/10.15252/embr.201845836
- Masetti, M. (2015, July 22). How many stars in the Milky Way? Retrieved from https://asd.gsfc.nasa.gov/blueshift/index.php/2015/07/22/how-many-stars-in-the-milky-way/